

Meeting Data files are now much smaller.Interface improved when creating the Midweek Meeting schedule and when selecting Persons for CLM parts.Life and Ministry Meeting Schedule: 2-per page Color Shaded.Timing of Treasures Talk and Spiritual Gems parts can now be adjusted.This allows very small languages to use NW Scheduler, and also allows brothers to manually add Meeting Data even without an EPUB file.
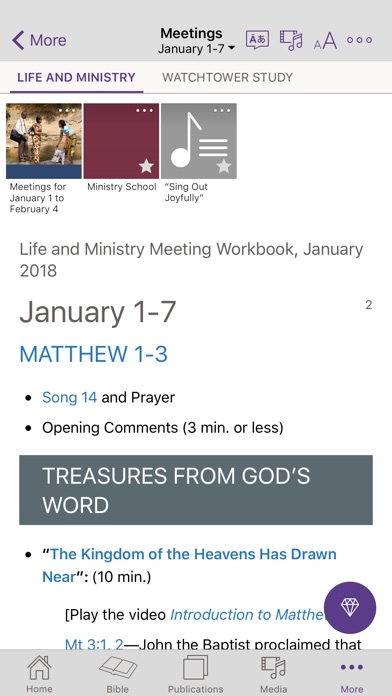
 All parts of the Life and Ministry Meeting can now be edited, e.g. talk titles, student parts, etc. Compare CLM Explorer with New World Scheduler. Compare Hourglass App with New World Scheduler. Compare Majestic KHS with New World Scheduler. New World Scheduler Reviews and Testimonials. sy2), Tripos Sybyl Line Notation (.sln), Beilstein ROSDAL (.ros), XYZ Files (. mmod), Schrödinger Maestro (.mae), Standard Molecular Data (.smd), Tripos Mol2 (.mol2. ent), RCSB Protein Data Bank Markup Language (.xml. mmcif), RCSB MacroMolecular Transmission Format (.mmtf), RCSB Protein Data Bank Files (.pdb. rd), MDL RXNFiles, both V2000 and V3000 connection tables (.rxn), MMI SketchEl Molecule (.el), Molinspiration JME String (.jme), RCSB Binar圜IF (.bcif), RCSB Macromolecular Crystallographic Information File (.cif. dx), ISIS Sketch File (.skc), ISIS Sketch Transportable Graphics File (.tgf), MDL MOLFiles, both V2000 and V3000 connection tables (.mol. com), IUPAC InChI (.inchi), IUPAC JCAMP-DX (.jdx. Read and write many popular chemical file types for working with the applications you use:ĪCD/ChemSketch Documents (.sk2), ChemDoodle Documents (.icl), ChemDoodle 3D Scenes (.ic3), ChemDoodle Javascript Data (.cwc.js), CambridgeSoft ChemDraw Exchange (.cdx), CambridgeSoft ChemDraw XML (.cdxml), Crystallographic Information Format (.cif), CHARMM CARD File (.crd), ChemAxon Marvin Document (.mrv), Chemical Markup Language (.cml), Daylight SMILES (.smi. Algorithmic Analysis of Cahn−Ingold−Prelog Rules of Stereochemistry: Proposals for Revised Rules and a Guide for Machine Implementation. and is 100% accurate in all 300 test cases provided. The CIP algorithm in ChemDoodle is validated against the test suite provided by Hanson et. Stereochemical features in your structures will be assigned "R", "S", "E", "Z", "M" and "P" descriptors. to remove any ambiguities and describe a completely consistent system for CIP assignments.ĬhemDoodle implements all 6 current CIP rules as well as auxilliary desciptors and mancude ring support. The most recent CIP rules from IUPAC were then algorithmically analyzed and standarized by Hanson et al. These rules were adopted by IUPAC for naming standards and fully described in the Blue books. While flawed, they have seen many revisions over the decades and were clarified by the work of Paulina Mata. The CIP rules have long been the standard for describing configurations of stereochemical features in a molecule.
All parts of the Life and Ministry Meeting can now be edited, e.g. talk titles, student parts, etc. Compare CLM Explorer with New World Scheduler. Compare Hourglass App with New World Scheduler. Compare Majestic KHS with New World Scheduler. New World Scheduler Reviews and Testimonials. sy2), Tripos Sybyl Line Notation (.sln), Beilstein ROSDAL (.ros), XYZ Files (. mmod), Schrödinger Maestro (.mae), Standard Molecular Data (.smd), Tripos Mol2 (.mol2. ent), RCSB Protein Data Bank Markup Language (.xml. mmcif), RCSB MacroMolecular Transmission Format (.mmtf), RCSB Protein Data Bank Files (.pdb. rd), MDL RXNFiles, both V2000 and V3000 connection tables (.rxn), MMI SketchEl Molecule (.el), Molinspiration JME String (.jme), RCSB Binar圜IF (.bcif), RCSB Macromolecular Crystallographic Information File (.cif. dx), ISIS Sketch File (.skc), ISIS Sketch Transportable Graphics File (.tgf), MDL MOLFiles, both V2000 and V3000 connection tables (.mol. com), IUPAC InChI (.inchi), IUPAC JCAMP-DX (.jdx. Read and write many popular chemical file types for working with the applications you use:ĪCD/ChemSketch Documents (.sk2), ChemDoodle Documents (.icl), ChemDoodle 3D Scenes (.ic3), ChemDoodle Javascript Data (.cwc.js), CambridgeSoft ChemDraw Exchange (.cdx), CambridgeSoft ChemDraw XML (.cdxml), Crystallographic Information Format (.cif), CHARMM CARD File (.crd), ChemAxon Marvin Document (.mrv), Chemical Markup Language (.cml), Daylight SMILES (.smi. Algorithmic Analysis of Cahn−Ingold−Prelog Rules of Stereochemistry: Proposals for Revised Rules and a Guide for Machine Implementation. and is 100% accurate in all 300 test cases provided. The CIP algorithm in ChemDoodle is validated against the test suite provided by Hanson et. Stereochemical features in your structures will be assigned "R", "S", "E", "Z", "M" and "P" descriptors. to remove any ambiguities and describe a completely consistent system for CIP assignments.ĬhemDoodle implements all 6 current CIP rules as well as auxilliary desciptors and mancude ring support. The most recent CIP rules from IUPAC were then algorithmically analyzed and standarized by Hanson et al. These rules were adopted by IUPAC for naming standards and fully described in the Blue books. While flawed, they have seen many revisions over the decades and were clarified by the work of Paulina Mata. The CIP rules have long been the standard for describing configurations of stereochemical features in a molecule.







 0 kommentar(er)
0 kommentar(er)
